Athena Starlard-Davenport1, Kristi Kutanzi2, Volodymyr Tryndyak2, Beverly Word1, Beverly Lyn-Cook1
1 Office of the Associate Director for Regulatory Activities, Jefferson, AR 72079, USA
2 Division of Biochemical Toxicology, FDA National Center for Toxicological Research, Jefferson, AR 72079, USA
| Date of Submission | 22-Aug-2012 |
| Date of Acceptance | 13-Dec-2013 |
| Date of Web Publication | 26-Jul-2013 |
Correspondence Address:
Athena Starlard-Davenport
Office of the Associate Director for Regulatory Activities, Jefferson, AR 72079
USA
Source of Support: None, Conflict of Interest: None
Abstract
It is well established that transcriptional silencing of critical tumor-suppressor genes by DNA methylation is a fundamental component in the initiation of breast cancer. However, the involvement of microRNAs (miRNAs) in restoring abnormal DNA methylation patterns in breast cancer is not well understood. Therefore, we investigated whether miRNA-29b, due to its complimentarity to the 3′- untranslated region of DNA methyltransferase 3A (DNMT3A) and DNMT3B, could restore normal DNA methylation patterns in human breast cancers and breast cancer cell lines. We demonstrated that transfection of pre-miRNA-29b into less aggressive MCF-7 cells, but not MDA-MB-231 mesenchymal cells, inhibited cell proliferation, decreased DNMT3A and DNMT3B messenger RNA (mRNA), and decreased promoter methylation status of ADAM23 , CCNA1, CCND2, CDH1, CDKN1C, CDKN2A, HIC1, RASSF1, SLIT2, TNFRSF10D, and TP73 tumor-suppressor genes. Using methylation polymerase chain reaction (PCR) arrays and real-time PCR, we also demonstrated that the methylation status of several critical tumor-suppressor genes increased as stage of breast disease increased, while miRNA-29b mRNA levels were significantly decreased in breast cancers versus normal breast. This increase in methylation status was accompanied by an increase in DNMT1 and DNMT3B mRNA in advanced stage of human breast cancers and in MCF-7, MDA-MB-361, HCC70, Hs-578T, and MDA-MB-231 breast cancer cells as compared to normal breast specimens and MCF-10-2A, a non-tumorigenic breast cell line, respectively. Our findings highlight the potential for a new epigenetic approach in improving breast cancer therapy by targeting DNMT3A and DNMT3B through miRNA-29b in non-invasive epithelial breast cancer cells.
Keywords: Breast cancer, DNA methylation, DNA methyltransferase1, DNA methyltransferases 3A, DNA methyltransferases 3B, microRNA-29b
How to cite this article:
Starlard-Davenport A, Kutanzi K, Tryndyak V, Word B, Lyn-Cook B. Restoration of the methylation status of hypermethylated gene promoters by microRNA-29b in human breast cancer: A novel epigenetic therapeutic approach. J Carcinog 2013;12:15
How to cite this URL:
Starlard-Davenport A, Kutanzi K, Tryndyak V, Word B, Lyn-Cook B. Restoration of the methylation status of hypermethylated gene promoters by microRNA-29b in human breast cancer: A novel epigenetic therapeutic approach. J Carcinog [serial online] 2013 [cited 2021 Oct 13];12:15. Available from: https://carcinogenesis.com/text.asp?2013/12/1/15/115720
Introduction
Breast cancer is the most prevalent malignancy and the second leading cause of cancer death in women. [1] Successful treatment of breast cancer relies on the ability to detect the disease early and correct molecular abnormalities associated with its development. Classic molecular cancer biology focuses on the role of direct DNA damage in the etiology of cancer; [2],[3],[4] however, it is now well documented that epigenetic alterations play a fundamental role in cancer initiation and progression, [5] including breast cancer. [6],[7],[8],[9] In addition, it is now well supported that altered gene-specific methylation triggered by abnormal functioning of DNA methyltransferases (DNMTs) may cause the transcriptional silencing of cancer-related genes in human cancers. [10],[11],[12] For instance, it has been demonstrated that DNMT1 and DNMT3B are overexpressed in breast cancer. [13] In support of the involvement of DNA methylation in transcriptional silencing, more than 100 individual tumor-suppressor genes, involved in multiple molecular pathways, have been identified to be frequently hypermethylated in breast cancer alone. [14] Recognition of the fundamental role of these epigenetic abnormalities in breast cancer progression [7],[14],[15],[16],[17] suggests that they could be used as biomarkers for clinical molecular diagnosis of breast cancer. [9],[18],[19],[20] However, the main question as to whether or not detection of these abnormal methylated genes can be used as therapeutic targets for breast cancer management, remains unresolved.
It is widely believed that reintroducing or reinforcing the expression of these epigenetically silenced genes is a rational strategy to restore control of critical cancer-related pathways and enhance the efficacy of cancer treatment. [21],[22],[23] As a result, the promising field of epigenetic therapy for cancer treatment has emerged. [24],[25],[26] Epigenetic modifiers, especially DNMT inhibitors such as 5-azacytidine and 5-aza-2′- deoxycytidine, are currently approved by the US Food and Drug Administration for the treatment of myelodysplastic syndrome. These drugs are incorporated into DNA during replication and their modified cytosine rings cause irreversible inactivation of DNMTs [22],[23] resulting in the re-expression of epigenetically silenced genes in vitro and in vivo. [25] However, treatment of solid tumors with epigenetic drugs thus far has been less promising compared to the treatment of myelodysplastic syndrome. [27],[28],[29],[30] The major obstacles in the modern field of epigenetic therapy include instability of epigenetic drugs in vivo, toxicity, and possible side effects associated with broad modes of action. In particular, results of recent studies have demonstrated a lack of association between clinical response and reversal of the methylated state of tumor-suppressor genes, suggesting a more complex mechanism behind their clinical efficacy may be involved. [25] This indicates the crucial need in developing more specific targeted epigenetic therapies.
Results of recent studies have suggested that cancer therapeutic strategies based on modulation of microRNA (miRNA) functioning in cancer cells hold great promise for cancer treatment. [31],[32],[33] Specifically, the enforced up-regulation of miRNA-29, [31] which is expressed in normal tissues but lost in cancers, caused inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and inhibition of tumorigenesis in vitro and in vivo. More importantly, studies by Fabbri et al., [31] and Garzon et al., [33] attributed the cancer inhibitory effect of miRNA-29b to its direct targeting of DNMTs DNMT3A and DNMT3B and suggested the use of miRNA-29b as a potentially effective hypomethylating agent. [33]
In this study, we initially investigated the methylation status of critical tumor-suppressor genes, expression of DNMTs, and miRNA-29b in human primary breast cancer tissues and breast cancer cell lines. Subsequently, the expression of miRNA-29b in MCF-7 and MDA-MB-231 human breast cancer cells was conducted by transfection of these cells with pre-miRNA-29b. Restoration of the abnormal methylation patterns of critical breast cancer-related genes in miRNA-29b-transfected MCF-7 cells was observed. This was evidenced by a substantial decrease in the extent of methylation of several hypermethylated tumor-suppressor genes; however, transfection of MDA-MB-231 cells with pre-miRNA-29b caused only moderate demethylation of five tumor-suppressor genes, namely CCNA1, CCND2, CDKN1C, RASSF1, and TP73 genes, while the methylation status of the majority of the genes remained either unaffected or slightly increased. In contrast, the demethylating effect of miRNA-29b in MDA-MB-231 cells was associated with down-regulation of the de novo DNMTs, DNMT3A and DNMT3B. Finally, these molecular epigenetic changes caused substantial inhibition of proliferation in miRNA-29b-transfected MCF-7 breast cancer cells, but not miRNA-29b-transfected MDA-MB-231 breast cancer cells. Collectively, our results highlight the potential for a new epigenetic approach in improving breast cancer therapy by targeting DNMT3A and DNMT3B through miRNA-29b in non-invasive epithelial breast cancer cells.
Materials and Methods
Cell lines and cell culture
Human MCF10-2A, MCF-7, MDA-MB-361, HCC70, BT-549, Hs-578T, and MDA-MB-231 breast cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained according to ATCC recommendations. Cells were seeded at a density of 0.5 × 10 6 viable cells per 100 mm plate, and the media were changed every other day for 6 days. Cells were harvested by incubation with 0.05% trypsin-Ethylenediaminetetraacetic acid (Invitrogen Corporation, Carlsbad, CA), washed in phosphate-buffered saline (PBS), and immediately frozen at -80°C for subsequent analyses.
Human breast cancer specimens
A total of 60 breast tissue specimens consisting of fresh frozen normal (n = 20), who primarily underwent bilateral mastectomy for reduction mammoplasty, and stage I-IV breast cancer (n = 40) specimens were purchased from the Cooperative Human Tissue Network (Birmingham, AL) and analyzed by quantitative real-time polymerase chain reaction (PCR). Samples were snap-frozen and stored at −80°C until analysis.
Of the 60 breast specimens obtained, 15 specimens were analyzed for gene-specific methylation using methylation PCR arrays (SABiosciences, Frederick, MD). Tissue specimens used for gene-specific methylation analysis consisted of five donors who had undergone reduction mammoplasty for macromastia (normals) and the remaining 10 donors had either surgical lumpectomies or mastectomies for the removal of the carcinoma. Patient age, tumor histologic type, tumor histologic grade, prognostic/predictive marker status, and history of neoadjuvant chemotherapy if available were retrospectively recorded from the medical records. Donor characteristics used in this study are listed in [Table 1]. All experiments that involved human tissue samples were reviewed and approved by the FDA Research Involving Human Subjects Committee and followed the principles embodied in the declaration of Helsinki.
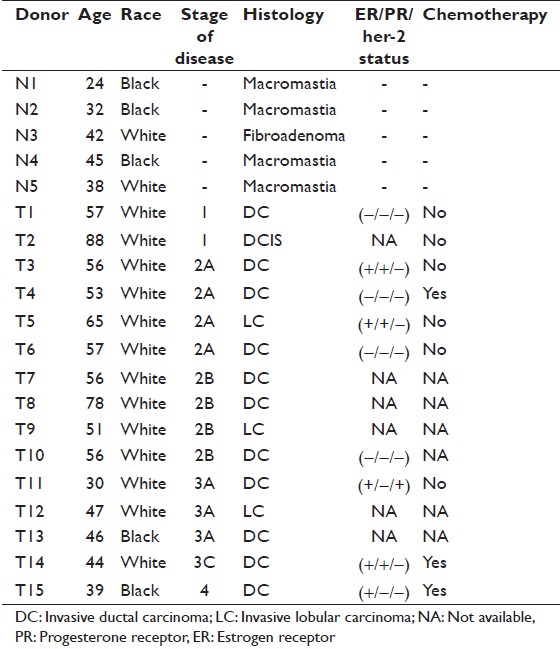 |
Table 1: Characteristics of donors analyzed in this study Click here to view |
RNA isolation and quantitative reverse transcription real-time polymerase chain reaction
Total RNA was extracted from breast cell lines and human breast tissue using TRI reagent (Ambion, Austin, TX) according to the manufacturer’s instructions. Reverse transcription was performed using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA).
The level of DNMT1, DNMT3A, and DNMT3B gene transcripts were determined by quantitative reverse transcription real-time (QRT-PCR) using TaqMan gene expression assays (Applied Biosystems) according to the manufacturer’s protocol. Relative quantification of gene expression was performed using glyceraldehyde-3-phosphate dehydrogenase as an internal control. QRT-PCR of miRNA-29b was performed using TaqMan miRNA assays (Applied Biosystems) according to the manufacturer’s instructions. RNU48 was used as endogenous control. The 2−ddCt method was used for calculating the relative amount of target RNA. [34] All QRT-PCR reactions were performed in triplicate, repeated at least 3 times, and always included a no-template sample as a negative control. Real-time PCR results are presented as average fold change of target gene in breast cancer cell lines relative to control, which gives the value 1.
Methylation polymerase chain reaction array
Genomic DNA was isolated from normal human breast tissue, breast cancer tissue, and MCF-7 and MDA-MB-231 breast cancer cell lines using DNeasy Blood and Tissue kits (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The methylation status of the tumor-suppressor genes most frequently hypermethylated in human breast cancer was determined using the Methyl-Profiler DNA Methylation PCR Array System (SABiosciences, Frederick, MD) according to the manufacturer’s protocol. A list of tumor-suppressor genes analyzed and their function are listed in [Table 2]. Each experiment was repeated twice, and each specimen was tested in triplicate.
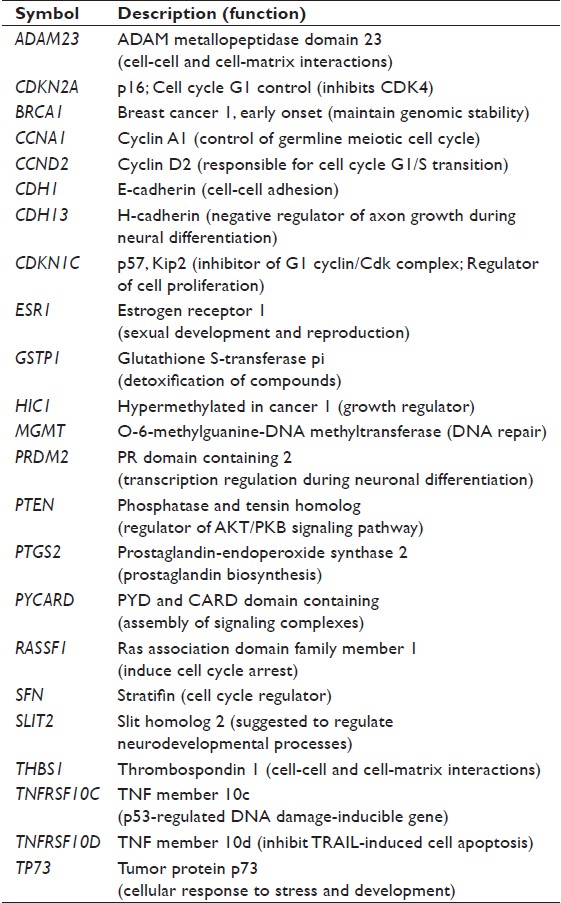 |
Table 2: Characteristics of tumor suppressor genes analyzed in this study Click here to view |
Transfection of breast cells with pre-microRNA-29b and analysis of cell growth
MCF-7 and MDA-MB-231 cells were seeded in 100 mm dishes at a density of 1 × 10 6 cells/dish, and transfected with 25 nM, 50 nM, and 100 nM of pre-miRNA-29b (Applied Biosystems) for 72 h in three independent replicates, using Lipofectamin™ 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. The cells transfected with scrambled RNA oligonucleotide served as control. The viability and growth of cells were monitored by the methyl thiazol tetrazolium test. Seventy-two hours after transfection, cells were harvested by incubation with 0.05% trypsin-EDTA (Invitrogen), washed in PBS, and immediately frozen at – 80°C for subsequent analyses.
Western blot analysis
Whole tissue and cell lysates were prepared by homogenization in 200 μl of lysis buffer (50 mM Tris-HCl, pH 7.4; 1% NP-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA; 1 mM PMSF; 1 μg/ml each aprotinin, leupeptin, pepstatin; 1 mM Na 3 VO 4 , 1 mM NaF), sonication, and incubation at 4°C for 30 min, followed by centrifugation at 12,000 × g at 4°C for 10 min. Extracts containing equal quantities of proteins were separated by SDS-PAGE on 8% polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were probed with primary antibodies against DNMT1 (1:1000; BD Biosciences, San Jose, CA), DNMT3A (1:200; Santa Cruz Biotechnology, Santa Cruz, CA), and DNMT3B (1:750; Millipore Corporation, Billerica, MA). Horseradish peroxidase-coupled bovine secondary antibodies (Santa Cruz Biotechnology) were used for visualization. Chemiluminescence detection was performed with the Immobilon Western Chemiluminescent HRP Substrate (Millipore Corporation) and measured directly by a BioSpectrum Imaging System (UVP, Upland, CA). Equal protein loading was confirmed by immunostaining against β-actin (1:4000; Sigma-Aldrich Corporation, St. Louis, MO). The signal intensity was analyzed by ImageQuant software (Molecular Dynamics, Sunnyvale, CA) and normalized to β-actin.
Statistical analysis
Statistical analyses were conducted by one-way analysis of variance, with pair-wise comparisons being conducted by Student-Newman-Keuls test. Results are presented as mean ± SD for analyzing the correlation between two categorical variables, Pearson’s coefficient (r) was used. All differences were considered statistically significant at the level of P < 0.05. All statistical analysis was performed using GraphPad Prism (version 4) software (GraphPad Software, Inc, La Jolla, CA).
Results
Methylation of tumor-suppressor genes in breast cancer
[Figure 1] shows the methylation status of 23 most frequently hypermethylated tumor-suppressor genes in human breast cancers (n = 15) and normal breast tissues (n = 5) samples as detected by the Methyl-Profiler DNA Methylation PCR Array System. Substantial DNA hypermethylation of tumor-suppressor genes was found in human breast tumor samples compared to normal (control) samples, including TP73, TNFRSF10C, HIC1, RASSF1A, CCND2, SLIT2, CDH13, CCNA1, and PTGS2 [Figure 1]. Interestingly, the frequency of hypermethylation of tumor-suppressor genes correlated with breast cancer progression. This was evidenced by a progressive increase in the number of hypermethylated genes at increasing stages of breast disease.
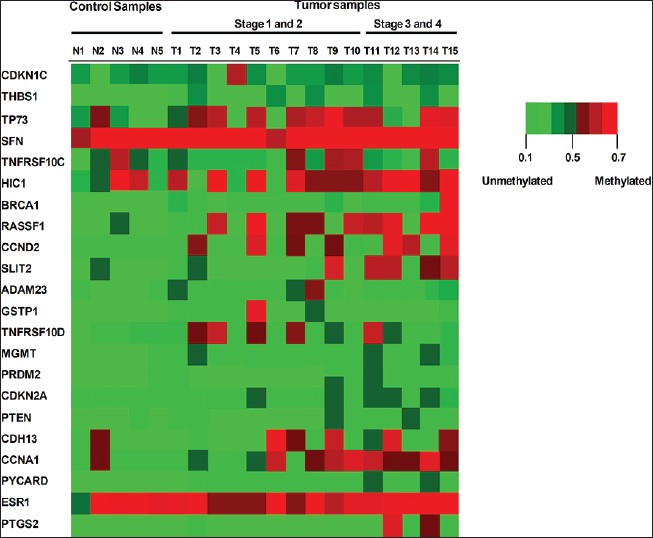 |
Figure 1: Gene-specific methylation changes in primary human breast cancers and normal (control) breast tissues. The methylation status of 24 tumor-suppressor genes, frequently hypermethylated in human primary breast cancer, was identified in 15 human breast tissues using the Methyl-Profiler DNA Methylation PCR Array System (SABiosciences). Samples N1-N5 represent control (normal) breast tissues samples and samples T1-T15 represent stages I-IV human breast tumor tissues from female donors. Each row represents a tumor-suppressor gene clone and each column represents a single DNA specimen. The degree of methylation is depicted by the level of intensity of the square, red representing greater than 25% promoter hypermethylation and green representing less than 20% promoter methylation (unmethylated) alleles for the tumor-suppressor gene clone, respectively Click here to view |
To determine whether the observed differences in methylation patterns were also evident in breast cell lines, we analyzed the methylation status of the same tumor-suppressor genes in MCF-7 and MDA-MB-231 breast cell lines [Figure 2]a. [Figure 2]a demonstrated that several genes, including ADAM23, CCND2, CDH13, CDKN2A, HIC1, RASSF1, and TP73, were hypermethylated in both MCF-7 and MDA-MB-231 cells. The mean percentage of hypermethylated tumor-suppressor genes was greater in the more aggressive MDA-MB-231 cells (32%) compared to the less aggressive MCF-7 cells (18%), a difference was significant (P = 0.02) (data not shown).
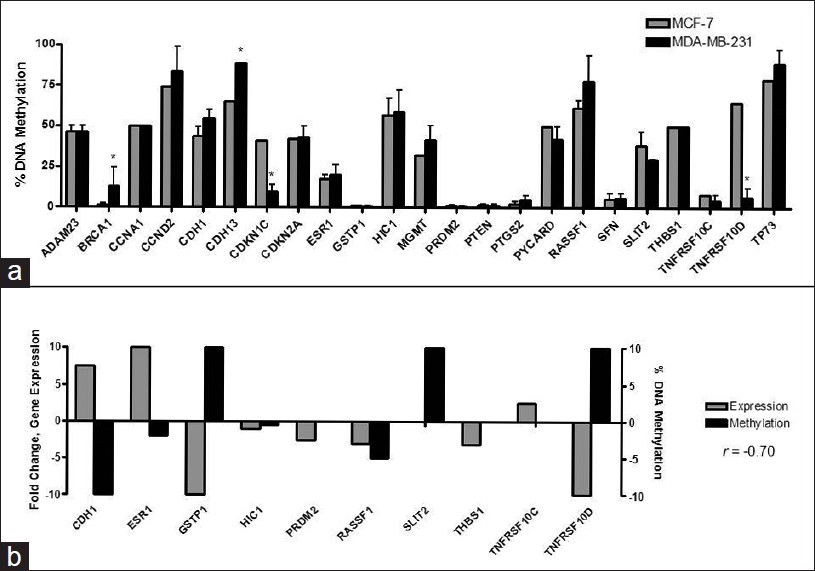 |
Figure 2: Evaluation of gene-specific methylation changes and gene expression in MCF-7 and MDA-MB-231 breast cell lines. (a) The methylation status of 24 tumor-suppressor genes, frequently hypermethylated in human primary breast cancer, was identified using the Methyl-Profiler DNA Methylation PCR Array System (SABiosciences). Grey bars (vertical) represent the percentage of DNA hypermethylation for a specif ic tumor-suppressor gene (horizontal) in MCF-7 cells and black bars (vertical) represent the percentage of DNA hypermethylation for a specific tumor-suppressor gene (horizontal) in MDA-MB-231 human breast cancer cells. Data are presented as percent of methylation ± SD. (b) Correlation of gene expression to gene – specific promoter methylation in MCF – 7 relative to MDA – MB – 231 breast cell lines Click here to view |
Because MDA-MB-231 breast cancer cells exhibit a more aggressive, basal-like mesenchymal phenotype than MCF-7, we were interested in determining whether hypermethylation of 10 tumor-suppressor genes from the Methylation PCR array correlated to a decrease in tumor-suppressor gene expression [Figure 2]b. Among the majority of tumor-suppressor genes, a decrease in gene expression correlated to an increase in DNA hypermethylation (Pearson’s correlation coefficient: r = −0.7) in both MCF-7 and MDA-MB-231 breast cancer cells. Specifically, this was evident among CDH1, ESR1, GSTP1, SLIT2, TNFRSF10C, and TNFRSF10D tumor-suppressor genes.
Expression of DNA methyltransferases DNMT1, DNMT3A, and DNMT3B in human breast tissues and breast cancer cell lines
The observed, aberrant gene-specific methylation changes in human breast cell lines prompted us to investigate whether or not these changes are associated with altered expression of DNMT1, DNMT3A, and DNMT3B in breast cancer tissues [Figure 3]a and breast cancer cell lines [Figure 3]b. We observed differential messenger RNA (mRNA) expression of DNMTs in breast cancers and normal human breast tissues. In human breast tissue specimens and stage I-IV breast cancers, we observed that DNMT1 and DNMT3B mRNA were significantly up-regulated in stages III-IV sporadic breast cancers as compared to adjacent normal tissue [Figure 3]a. Although DNMT3A mRNA was slightly increased in stage III-IV breast cancers as compared to adjacent normal breast tissues, its expression in stage III-IV breast cancers as compared to adjacent normal breast tissues was not significantly up-regulated (P = 0.157). Furthermore, there was no difference in DNMT3A mRNA expression levels in stage I-II breast cancers compared to adjacent normal breast tissues (P = 0.548).
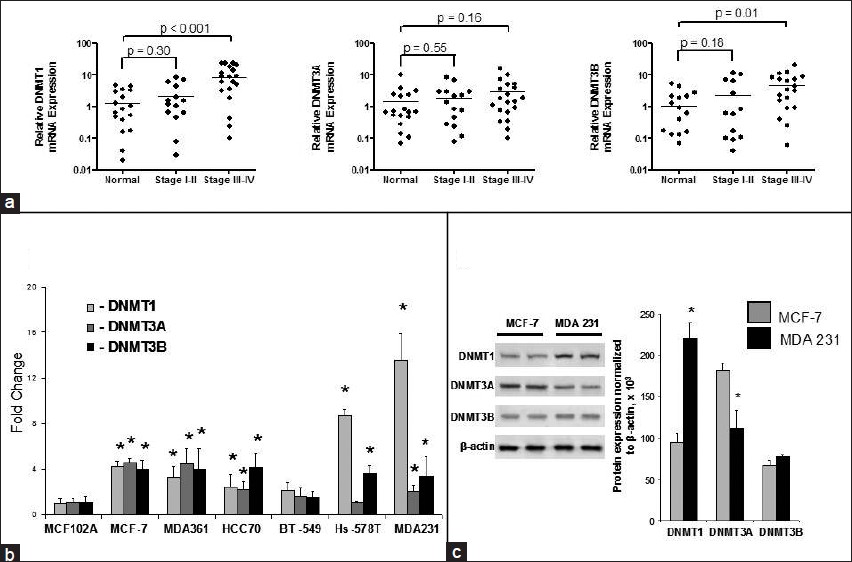 |
Figure 3: Expression of DNMT1, DNMT3A, and DNMT3B in human breast tissues and breast cell lines. (a) DNMT1, DNMT3A, and DNMT3B mRNA expression in adjacent normal breast tissues and stages I-IV breast cancers. (b) DNMT1, DNMT3A, and DNMT3B mRNA expression in six breast cancer cell lines and MCF-10-2A, a non-tumorigenic breast cell line. (c) DNMT1, DNMT3A, and DNMT3B protein expressions in MCF-10-2A, MCF-7, and MDA-MB-231 breast cell lines. *Indicates significant difference from MCF-10-2A non-tumorigenic breast cell line Click here to view |
In a panel of six breast cancer cell lines and the normal MCF-10-2A breast cell line, all three functional DNMTs DNMT1, DNMT3A, and DNMT3B were expressed at higher levels in MCF-7, MDA-MB-361, HCC70, and MDA-MB-231 human breast cancer cell lines than in non-tumorigenic MCF-10-2A cells [Figure 3]b. However, each breast cancer cell line was characterized by different expression patterns within DNMTs. Specifically, MCF-7 cells were characterized by a 4-fold up-regulation of DNMT1, DNMT3A, and DNMT3B as compared to non-tumorigenic MCF-10-2A cells, whereas Hs-578T and MDA-MB-231 cells exhibited a profound up-regulation of DNMT3B. The level of DNMT1 mRNA expression in the more aggressive Hs-578T and MDA-MB-231 invasive mesenchymal breast cancer cells was 8 and 13 times greater, respectively, as compared to MCF-10-2A and MCF-7 cells. This up-regulation of DNMT mRNA in MCF-7 and MDA-MB-231 breast cancer cells compared to MCF-10-2A was further confirmed by Western blot analysis [Figure 3]c.
Expression of microRNA-29b in breast cancer
In order to determine the mechanism associated with altered DNMT expression in breast cancers, we searched for a link between DNMT3A and DNMT3B expression and altered miRNA expression. Interestingly, a member of the miR-29 family, miRNA-29b, has a sequence complementarity to the sequence in the 3′- untranslated region (3′- UTR) of the human DNMT3A and DNMT3B gene ( www.targetscan.org ) [Figure 4]a. [Figure 4]b shows significant down-regulation of miRNA-29b expression, a known-regulator of DNMT3A and DNMT3B, [31],[33] in breast cancer tissues. Similar to primary breast tissue, MCF-7 cells exhibited substantial down-regulation of miRNA-29b expression as compared to non-tumorigenic MCF-10-2A cells [Figure 4]c. Surprisingly, we found a significant up-regulation of miRNA-29b in MDA-MB-231 cells [Figure 4]c.
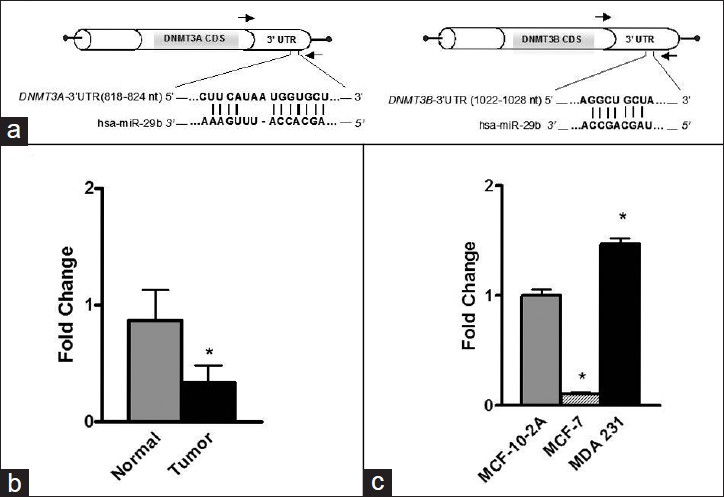 |
Figure 4: miRNA – 29b directly targets DNMT3A and DNMT3B. (a) Complementary sequence for miRNA – 29b in the 3 – UTR of DNMT3A and DNMT3B. (b) Expression of miRNA – 29b in normal and tumor tissues. (c) Expression of miRNA – 29b in MCF – 10 – 2A, MCF – 7, and MDA – MB – 231 breast cell lines Click here to view |
Effect of ectopic up-regulation of miRNA-29b on the methylation status of tumor-suppressor genes in MCF-7 and MDA-MB-231 cells
To determine whether or not expression of miRNA-29b would affect gene-specific methylation and inhibit breast cancer cell growth in MDA-MB-231 cells, we transfected MDA-MB-231 cells with pre-miRNA-29b. [Figure 5]a shows that transfection with pre-miRNA-29b for 72 h substantially elevated miRNA-29b levels [Figure 5]a, which was associated with a decrease in expression of DNMT3A and DNMT3B by 60% and 44%, respectively [Figure 5]b. In contrast, the expression of DNMT1 in miRNA-29b/MDA-MB-231-transfected cells did not change [Figure 5]b. These changes in the expression of DNMT3A and DNMT3B resulted in moderate demethylation of CCNA1, CCND2, CDKN1C, RASSF1A, SFN, and TP73 genes only, while the methylation status of the majority of genes either increased or remained unaffected [Figure 5]c. In contrast, 72-h transfection of MCF-7 cells with pre-miRNA-29b resulted in a substantial decrease in the extent of methylation in the majority of hypermethylated tumor-suppressor genes, including ADAM23, CCNA1, CCND2, CDH1, CDKN1C, CDKN2A, HIC1, MGMT, RASSF1, SFN, SLIT2, TNFRSF10C, TNFRSF10D, and TP73 [Figure 6]a. These demethylation changes were associated with inhibition of cell growth at 50 and 100 nM concentrations of pre-miRNA-29b in MCF-7 cells as compared to control (0 nM) cells [Figure 6]b. In contrast, 72-h transfection with 0, 25, 50, or 100 nM of pre-miRNA-29b did not inhibit cell growth of MDA-MB-231 cells [Figure 6]c.
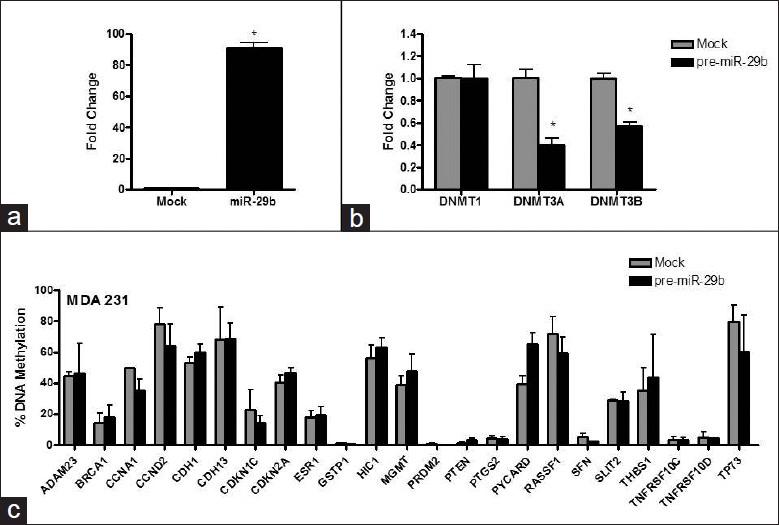 |
Figure 5: Effect of ectopic up-regulation of miRNA-29b in MDA-MB-231 cells. (a) miRNA-29b expression in MDA-MB-231 cells after 72 h transfection with pre – miR – 29b (MDA-MB-231/ miRNA-29b) and with scrambled RNA oligonucleotide (mock) served as a control (0 nM pre-miRNA-29b). (b) DNMT1, DNMT3A, and DNMT3B mRNA expressions in mock (control) and pre-miRNA-29b-transfected MDA-MB-231cells. (c) Gene-specific methylation analysis after transfection of MDA-MB-231 cells with pre-miRNA-29b Click here to view |
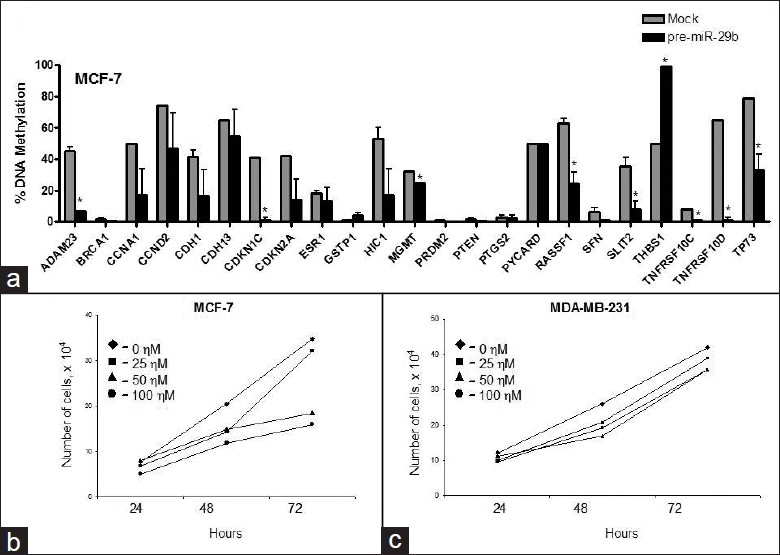 |
Figure 6: Effect of ectopic up-regulation of miRNA-29b in MCF-7 cells. (a) Gene-specific methylation analysis after transfection of MCF-7 cells with pre-miRNA-29b. (b) Cell viability assay of 0, 25, 50, and 100 nM pre-miRNA-29b in MCF-7/ miRNA-29b cells after 24, 48, and 72 h. (c) Cell viability assay of 0, 25, 50, and 100 nM pre-miRNA-29b in MDA-MB-231/miRNA-29b cells after 24, 48, and 72 h Click here to view |
Discussion
Aberrant DNA methylation is an important epigenetic event that has been found to be associated with tumorigenesis. [9] Accumulated evidence during recent years has established that hypermethylation of cytosine phosphate guanine (CpG) island-containing promoters of critical cancer-related genes and the concomitant transcriptional inhibition of gene expression are the most frequent epigenetic abnormalities found in all major human cancers, [5] including breast cancer. [7],[9] In our study, we analyzed the methylation status of 23 tumor-suppressor genes in breast tissues and MCF-7 and MDA-MB-231 breast cell lines. Similar to previous more comprehensive reports, [9],[10],[11] we observed progressive tumor-suppressor gene-specific hypermethylation in both human breast cancer tissues and breast cancer cell lines [Figure 1] and [Figure 2]a. Specifically, we found substantial hypermethylation of several analogous tumor-suppressor genes in human breast tumor samples including CCNA1, CCND2, CDH13, RASSF1A, SLIT2, TNFRSF10C, and TP73. Numerous studies have linked this gene-specific hypermethylating event in breast cancer to the increased activity, expression, or protein stability of one or more of the three known catalytically active DNMTs [14],[35],[36] which were analyzed in this study. We observed an increase in gene expression of DNMT1 and DNMT3B in more aggressive breast cancer cells of basal-like mesenchymal phenotype, MDA-MB-231 and Hs-578T, and advanced stage of breast disease. In support of our research findings, a similar study demonstrated that DNMT3B mRNA, not DNMT3A, levels were highly expressed in breast tumors. [13] In addition, this finding corresponds to the suggestion that overexpression of DNMT1 and DNMT3B contributes to the establishment of a “hypermethylator” phenotype in invasive human mesenchymal breast cancer cells, [36] and therefore may be a crucial biomarker for predicting more aggressive and advanced stage of breast carcinogenesis. However, the mechanism responsible for cancer-specific DNMTs up-regulation is largely unknown.
Several groups of investigators have reported a crucial role of miRNAs s in control of DNA methylation through either direct targeting of DNMT3A and DNMT3B by miRNA-29b. [31],[33],[37] In this study, we found that in human breast cancer tissues, up-regulation of DNMT3B was accompanied by an overall decrease in expression of miRNA-29b. To our knowledge, this is the first report that provides evidence of miRNA-29b down-regulation in human breast cancer tissues and MCF-7 breast cancer cells. The finding that up-regulation of DNMT3B is associated with a decrease in miRNA-29b in breast tissues and breast cancer cells suggests that miRNA-29b, similarly to its role in lung cancer and acute myeloid leukemia, [33] may control the functioning of DNMT3B in breast cancer. Therefore, loss of miRNA-29b in breast carcinomas may lead to overexpression of DNMT3B and concomitant hypermethylation of critical tumor-suppresor genes.
Results of recent studies conducted by Fabbri et al., [31] and Garzon et al., [33] demonstrated that enforced ectopic expression of miRNA-29b inhibits progression of lung cancer in mice and partially diminishes tumor-specific phenotype in acute myeloid leukemia Kasumi-1 cells. In this study, we demonstrate that the methylation status of the majority of tumor-suppressor genes analyzed in this study was significantly decreased after restoration of miRNA-29b expression in MCF-7 epithelial breast cancer cells and cell growth decreased. On the other hand, enforced ectopic up-regulation of miRNA-29b in MDA-MB-231 breast cancer cells did not affect the methylation status of tumor-suppressor genes or prevent tumor cell growth in invasive MDA-MB-231 mesenchymal breast cancer cells. Despite previous finding showing that miRNA-29b can indirectly down-regulate DNMT1 by targeting zinc finger transcription factor Sp1, [33] in this study we did not detect changes in DNMT1 expression neither in miRNA-29b-transfected MCF-7 nor MDA-MB-231 cells. This discrepancy may be more attributed to Sp3-dependent up-regulation of DNMT1 in MDA-MB-231 cells, rather than Sp1-dependent. This is evidenced by a survey analysis of a panel of breast cancer cell lines showing that MDA-MB-231 cells are characterized by a greater expression of Sp3 transcription factor and lower expression of Sp1. [38] Therefore, DNMT1 in MDA-MB-231 cells may be targeted through Sp3, not Sp1 transcription factor.
Collectively, results from our study suggest that restoration of the methylation status of hypermethylated gene promoters by targeting DNMT3B through miRNA-29b in less aggressive epithelial breast cancers could be a promising avenue for cancer therapy.
Acknowledgments
This work was supported by a generous grant from the US Food and Drug Administration Office of Women’s Health to AS-D.
References
| 1. | DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin 2011;61:409-18  |
| 2. | Srivastava S, Grizzle WE. Biomarkers and the genetics of early neoplastic lesions. Cancer Biomark 2010;9:41-64  |
| 3. | Lambrechts S, Decloedt J, Neven P. Breast cancer prevention: Lifestyle changes and chemoprevention. Acta Clin Belg 2011;66:283-92.  [PUBMED] |
| 4. | Bell DW. Our changing view of the genomic landscape of cancer. J Pathol 2010;220:231-43.  [PUBMED] |
| 5. | Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92.  [PUBMED] |
| 6. | Polyak K. Breast cancer: Origins and evolution. J Clin Invest 2007;117:3155-63.  [PUBMED] |
| 7. | Veeck J, Esteller M. Breast cancer epigenetics: From DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia 2010;15:5-17.  [PUBMED] |
| 8. | Dalvai M, Bystricky K. The role of histone modifications and variants in regulating gene expression in breast cancer. J Mammary Gland Biol Neoplasia 2010;15:19-33.  [PUBMED] |
| 9. | Martens JW, Margossian AL, Schmitt M, Foekens J, Harbeck N. DNA methylation as a biomarker in breast cancer. Future Oncol 2009;5:1245-56.  [PUBMED] |
| 10. | Vilain A, Vogt N, Dutrillaux B, Malfoy B. DNA methylation and chromosome instability in breast cancer cell lines. FEBS Lett 1999;460:231-4.  [PUBMED] |
| 11. | Chimonidou M, Strati A, Tzitzira A, Sotiropoulou G, Malamos N, Georgoulias V, et al. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem 2011;57:1169-77.  |
| 12. | Esteller M. Epigenetic lesions causing genetic lesions in human cancer: Promoter hypermethylation of DNA repair genes. Eur J Cancer 2000;36:2294-300.  [PUBMED] |
| 13. | Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res 2003;9:4415-22.  |
| 14. | Hinshelwood RA, Clark SJ. Breast cancer epigenetics: Normal human mammary epithelial cells as a model system. J Mol Med (Berl) 2008;86:1315-28.  [PUBMED] |
| 15. | Radpour R, Kohler C, Haghighi MM, Fan AX, Holzgreve W, Zhong XY. Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass array. Oncogene 2009;28:2969-78.  [PUBMED] |
| 16. | Brooks J, Cairns P, Zeleniuch-Jacquotte A. Promoter methylation and the detection of breast cancer. Cancer Causes Control 2009;20:1539-50.  [PUBMED] |
| 17. | Jovanovic J, Rønneberg JA, Tost J, Kristensen V. The epigenetics of breast cancer. Mol Oncol 2010;4:242-54.  |
| 18. | Levenson VV. Biomarkers for early detection of breast cancer: What, when, and where? Biochim Biophys Acta 2007;1770:847-56.  [PUBMED] |
| 19. | Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol 2009;19:165-71.  [PUBMED] |
| 20. | Suijkerbuijk KP, van Diest PJ, van der Wall E. Improving early breast cancer detection: Focus on methylation. Ann Oncol 2011;22:24-9.  [PUBMED] |
| 21. | Esteller M. DNA methylation and cancer therapy: New developments and expectations. Curr Opin Oncol 2005;17:55-60.  [PUBMED] |
| 22. | Momparler RL. Epigenetic therapy of cancer with 5-aza-2′- deoxycytidine (decitabine). Semin Oncol 2005;32:443-51.  [PUBMED] |
| 23. | Cortez CC, Jones PA. Chromatin, cancer and drug therapies. Mutat Res 2008;647:44-51.  [PUBMED] |
| 24. | Kristensen LS, Nielsen HM, Hansen LL. Epigenetics and cancer treatment. Eur J Pharmacol 2009;625:131-42.  [PUBMED] |
| 25. | Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem 2009;16:2075-85.  [PUBMED] |
| 26. | Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res 2009;15:3938-46.  [PUBMED] |
| 27. | de Vos D. Epigenetic drugs: A longstanding story. Semin Oncol 2005;32:437-42.  [PUBMED] |
| 28. | Ghoshal K, Bai S. DNA methyltransferases as targets for cancer therapy. Drugs Today (Barc) 2007;43:395-422.  [PUBMED] |
| 29. | Graham JS, Kaye SB, Brown R. The promises and pitfalls of epigenetic therapies in solid tumours. Eur J Cancer 2009;45:1129-36.  [PUBMED] |
| 30. | Daskalakis M, Blagitko-Dorfs N, Hackanson B. Decitabine. Recent Results Cancer Res 2010;184:131-57.  [PUBMED] |
| 31. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 2007;104:15805-10.  |
| 32. | Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137:1005-17.  |
| 33. | Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009;113:6411-8.  |
| 34. | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101-8.  [PUBMED] |
| 35. | Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem 2005;280:18302-10.  |
| 36. | Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer 2008;7:15.  [PUBMED] |
| 37. | Takada S, Berezikov E, Choi YL, Yamashita Y, Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA 2009;15:1507-14.  [PUBMED] |
| 38. | Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 2007;67:11001-11.  [PUBMED] |
Authors
Dr. Athena Starlard-Davenport, Office of the Associate Director for Regulatory Activities, National Center for Toxicological Research, Jefferson, AR, 72079, USA.
Dr. Kristy Kutanzi, Department of Biochemical Toxicology, National Center for Toxicological Research, Jefferson, AR 72079, USA.
Dr. Volodymyr Tryndyak, Department of Biochemical Toxicology, National Center for Toxicological Research, Jefferson, AR 72079, USA.
Miss. Beverly Word, Office of the Associate Director for Regulatory Activities, National Center for Toxicological Research, Jefferson, AR 72079, USA.
Dr. Beverly Lyn-Cook, Office of the Associate Director for Regulatory Activities, National Center for Toxicological Research, Jefferson, AR 72079, USA.
