Kathleen A Vermeersch, Mark P Styczynski
School of Chemical & Biomolecular Engineering and Institute for Bioengineering & Bioscience, Georgia Institute of Technology, 311 Ferst Dr. NW, Atlanta, GA 30332-0100, USA
| Date of Submission | 22-Feb-2013 |
| Date of Acceptance | 12-May-2013 |
| Date of Web Publication | 18-Jun-2013 |
Correspondence Address:
Mark P Styczynski
School of Chemical & Biomolecular Engineering and Institute for Bioengineering & Bioscience, Georgia Institute of Technology, 311 Ferst Dr. NW, Atlanta, GA 30332-0100
USA
Source of Support: DARPA grant YFA N66001-11-1-4128, NIH grant R21 CA167500 and the Integrative Bio Systems Institute of Georgia Tech., Conflict of Interest: None
DOI: 10.4103/1477-3163.113622
Abstract
The first discovery of metabolic changes in cancer occurred almost a century ago. While the genetic underpinnings of cancer have dominated its study since then, altered metabolism has recently been acknowledged as a key hallmark of cancer and metabolism-focused research has received renewed attention. The emerging field of metabolomics – which attempts to profile all metabolites within a cell or biological system – is now being used to analyze cancer metabolism on a system-wide scale, painting a broad picture of the altered pathways and their interactions with each other. While a large fraction of cancer metabolomics research is focused on finding diagnostic biomarkers, metabolomics is also being used to obtain more fundamental mechanistic insight into cancer and carcinogenesis. Applications of metabolomics are also emerging in areas such as tumor staging and assessment of treatment efficacy. This review summarizes contributions that metabolomics has made in cancer research and presents the current challenges and potential future directions within the field.
Keywords: Biomarkers, carcinogenesis, metabolomics
How to cite this article:
Vermeersch KA, Styczynski MP. Applications of metabolomics in cancer research. J Carcinog 2013;12:9
How to cite this URL:
Vermeersch KA, Styczynski MP. Applications of metabolomics in cancer research. J Carcinog [serial online] 2013 [cited 2021 Oct 13];12:9. Available from: https://carcinogenesis.com/text.asp?2013/12/1/9/113622
Introduction
Broadly defined, metabolism is the set of processes catalyzing the production of energy and cellular building blocks (amino acids, nucleotides, lipids, etc.) from the nutrients a cell takes up from the environment. These building blocks and the biochemical intermediates generated during their production and utilization are collectively referred to as metabolites. Metabolite levels integrate the effects of gene regulation, post-transcriptional regulation, pathway interactions, and environmental perturbations; this downstream synthesis of diverse signals ultimately makes metabolites direct molecular readouts of cell status that reflect a meaningful physiological phenotype. [1],[2],[3],[4] Metabolomics, then, is the emerging field focused on comprehensive profiling of metabolites in a sample, whether intracellular or from circulating biofluids. The ability of metabolomics to measure high-throughput, system-wide phenotypes gives it great power in the field of oncology to further understand what is happening in cancer cells. In this review, we will focus on specific areas within cancer and carcinogenesis research on which metabolomics has been brought to bear.
Common Metabolic Alterations in Cancer
Though reprogramming of energy metabolism was only recently recognized as an emerging hallmark of cancer, [5] altered cancer metabolism was first identified almost a century ago when Warburg discovered that cancer cells primarily use anaerobic glycolysis to produce their energy instead of oxidative phosphorylation, even in the presence of oxygen – a phenomenon known as the Warburg effect or aerobic glycolysis. [6],[7] Over the years, many common cancer mutations have been shown to support the Warburg effect. [8] AKT1 (a serine/threonine kinase), hypoxic inducing factor (HIF), and p53 (a tumor suppressor protein) together cause increased flux of glucose through glycolysis and down-regulation of flux through the tricarboxylic acid (TCA) cycle [Figure 1], thereby supporting the Warburg effect and carcinogenesis. [9],[10],[11],[12],[13],[14],[15],[16],[17] Loss-of-function mutations of mitochondrial enzymes succinate dehydrogenase (SDH) and fumarate hydratase (FH) also support the Warburg effect via accumulation of succinate and fumarate – metabolites that inhibit prolyl hydroxylases (PHD), a family of enzymes that tag HIF for degradation. [18],[19],[20],[21],[22] Thus, changes in SDH and FH support HIF accumulation, which in turn supports carcinogenesis and the Warburg effect.
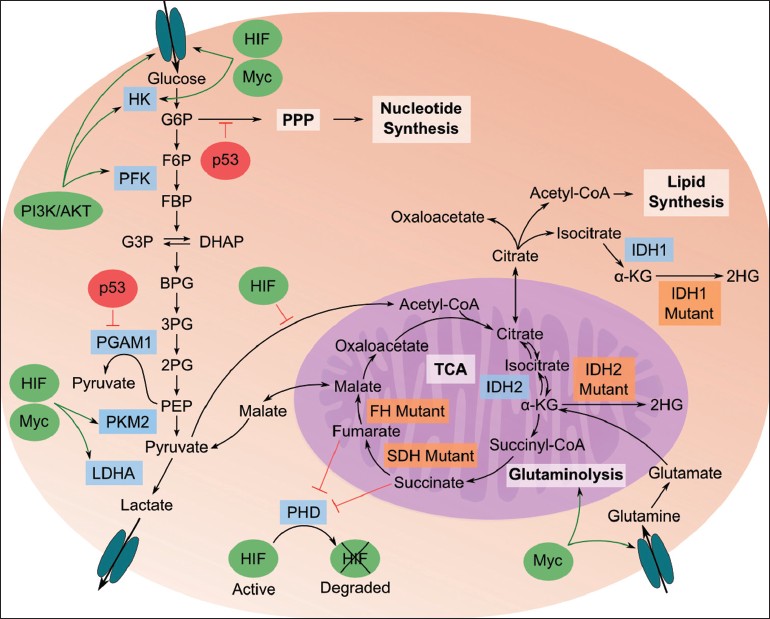 |
Figure 1: Illustration of important relationships between metabolome, proteome, and genome in cancerous cells. Glycolysis breaks down glucose into pyruvate, which is then fermented to lactate; pyruvate flux through the tricarboxylic acid (TCA) cycle is down-regulated in cancer cells. Pathways branching off of glycolysis, such as the pentose phosphate pathway (PPP), generate biochemical building blocks to sustain the high proliferative rate of cancer cells. Specific genetic and enzyme-level behaviors are described in the main text. Blue boxes are enzymes important in transitioning to a cancer metabolic phenotype; orange boxes are enzymes that are mutated in cancer cells. Green ovals are oncogenes that are up-regulated in cancer; red ovals are tumor suppressors that are down-regulated in cancer. Figure abbreviations: 2PG: 2-phosphoglycerate; 3PG: 3-phosphoglycerate; BPG: 1,3-bisphosphoglycerate; CoA: coenzyme A; DHAP: dihydroxyacetone phosphate; F6P: fructose-6-phosphate; FBP: fructose-1,6-bisphosphate; G3P: glyceraldehyde-3-phospate; G6P: glucose-6-phosphate; HK: hexokinase; LDHA: lactate dehydrogenase A; PFK: phosphofructokinase; PI3K: phosphatidylinositide 3-kinase Click here to view |
Another important altered pathway in cancer metabolism is glutaminolysis, a key source of energy and anaplerotic precursors for the TCA cycle. [23] Myc, an oncogenic transcription factor, interacts with HIF to regulate several enzymes in glucose metabolism and plays an important role in glutaminolysis [Figure 1]. [24],[25] Besides stimulating the glutamine transporter, Myc indirectly regulates glutaminase (GLS), a mitochondrial enzyme that converts glutamine to glutamate, through transcriptional repression of the microRNAs that repress GLS. [26]
Pyruvate kinase (PK) is another common cancer signature with metabolic implications [Figure 1]. PK catalyzes phosphoenolpyruvate (PEP) conversion into pyruvate, a rate-limiting step in glycolysis. It is widely believed that during carcinogenesis, there is a change in expression of PK isoforms, from pyruvate kinase isozyme M1 towards less-active, rate-limiting pyruvate kinase isozyme M2 (PKM2), [27] potentially leading to accumulation of upstream glycolytic intermediates [8] . Additional changes in cancer metabolism prevent such accumulation (which would lead to down-regulation of glycolysis) by channeling these intermediates into branching pathways, which produces higher levels of these pathways’ end products such as reduced nicotinamide adenine dinucleotide phosphate (NADPH), ribose 5-phosphate, and nucleic acids. [28],[29]
In addition, an alternative pathway for pyruvate fermentation has recently been found that converts PEP to pyruvate through the direct phosphorylation of phosphoglycerate mutase 1 (PGAM1) without production of adenosine-5′- triphosphate (ATP) [Figure 1]. [30] By decoupling energy generation from glycolysis, pyruvate production from PEP continues regardless of ATP regulation or dependence on PKM2. This continuous pyruvate production, along with glutaminolysis, accounts for the characteristically high levels of lactic acid in tumors.
Metabolomics and Cancer
Analytical technology
Currently, no single analytical method can measure concentrations of all metabolites due to their significant chemical diversity. The two dominant metabolomics technologies are nuclear magnetic resonance (NMR) and mass spectrometry (MS) coupled to a separation technique. Both of these technologies and the roles they play in metabolomics are extensively detailed elsewhere, [31],[32],[33] but a brief description will be given here.
NMR
NMR provides quantitative and structural information and can measure a wide range of metabolites with little to no sample preparation. One limitation of NMR is its low sensitivity and thus higher limits of detection for metabolites. Additionally, in complex mixtures the interpretability of NMR spectra and association to specific metabolite identities can be difficult. Techniques including high-resolution NMR and high-resolution magic angle spinning NMR (HR-MAS-NMR) have been used to profile cancer metabolism in biofluids as well as tissue samples; they are particularly valuable since they do not destroy samples, allowing for parallel analysis with other techniques. [34],[35] Another emerging technology, hyperpolarized NMR, has been used to characterize cancer metabolism by tracing metabolite levels in vivo,[36] with potential applications in clinical diagnosis or treatment of cancer. [37]
MS
MS provides semi-quantitative information with very high sensitivity, allowing the analysis of low-abundance metabolites. Many MS-based techniques require extensive sample preparation and usually can only measure specific subsets of metabolites. MS-based analyses can be broadly divided into direct injection techniques – including direct infusion MS [38] and direct analysis in real time MS (DART-MS) [39] – and separation-coupled techniques, including gas chromatography-MS (GC-MS), liquid chromatography-MS (LC-MS) and capillary electrophoresis-MS (CE-MS). Common types of mass spectrometers include time-of-flight (TOF), quadrupole time-of-flight (QTOF), quadrupole, and orbitrap. Separation methods and MS can also be combined in series (GCxGC-MS or LC-MS/MS) to gain better separation or more structural information.
Data handling and processing
The complex raw data captured by metabolomics instruments must first be converted into human-interpretable measurements; the resulting vast datasets then require significant analysis and interpretation. Numerous data processing techniques and packages have been created for all steps of this data-processing pipeline. We refer the reader to in-depth discussions of available bioinformatics tools elsewhere. [40]
Study of carcinogenesis and cancer biology
The system-wide analyses of metabolomics allow a unique opportunity for the study of carcinogenesis and cancer biology by enabling deep investigation of targeted aspects of cancer metabolism while also allowing discovery-based analysis of metabolism writ large.
For example, nicotinamide adenine dinucleotide phosphate (NADP + )-dependent isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) are commonly subject to gain-of-function point mutations in gliomas. [41] Using metabolomics, it was discovered that mutated IDH1 and IDH2 catalyze (R)-2-hydroxyglutarate (2HG), a rare metabolite, from α-ketoglutarate (α-KG) [Figure 1]. [42] 2HG has been referred to as an oncometabolite because its production helps to further the cancer phenotype. [42],[43] Metabolic profiling on glioma cells using LC-MS/MS and GC-MS showed that IDH1/2 mutations caused N-acetylated amino acids and TCA cycle intermediate levels to drop and biosynthetic molecules to accumulate while not affecting glycolytic intermediates. [44] The effects of IDH1/2 mutations on the metabolome were very similar to the changes caused by treating normal cells with 2HG, showing that it is the production of the oncometabolite 2HG and not the loss of IDH1/2’s normal operation that causes these changes. [44]
Another (though somewhat disputed [45] ) example of mechanistic insight from metabolomics is in sarcosine’s putative role in prostate cancer progression. Samples from patients with benign, localized, and metastatic prostate cancer were profiled using both LC-MS and GC-MS. From this metabolic profiling, sarcosine levels were identified as increasing from benign to metastatic prostate cancer. In vitro, sarcosine levels were shown to directly correlate to a cell’s level of invasiveness. Further investigation showed that sarcosine is regulated by an androgen receptor and ETS gene family fusions through transcriptional control of its regulatory enzymes. [46]
A final example of metabolomics-based mechanistic insight is the recent study of extracellular metabolite profiles across the National Cancer Institute’s NCI-60 cancer cell lines. [47] Glycine consumption was found to be correlated with proliferation rate in cancerous cells, but not in proliferative non-cancerous cells, suggesting cancer-specific behavior. De novo purine nucleotide biosynthesis was one pathway involved in the increased glycine consumption. Follow-up analysis of breast cancer gene expression data revealed that glycine mitochondrial enzyme expression correlated with cancer mortality.
Biomarkers and diagnosis
A central focus in cancer metabolomics research is biomarker discovery. Metabolites are theoretically ideal biomarkers and diagnostics because they can be easily measured from non-invasive urine or blood samples. Many groups are attempting to use metabolic profiles as biomarkers or diagnostic tools, for essentially every type of cancer, since levels of multiple metabolites can provide better classification than a single entity. For example, the diagnostic capability of a set of 113 cis?diol structured urinary metabolites for liver cancer resulted in a lower false?positive rate than the traditional tumor marker alpha?fetoprotein when classifying liver cancer against hepatocirrhosis and chronic hepatitis samples-diol structured urinary metabolites for liver cancer resulted in a lower false-positive rate than the traditional tumor marker alpha-fetoprotein when classifying liver cancer against hepatocirrhosis and chronic hepatitis samples. [48] A representative, but necessarily incomplete, selection of applications of metabolomics for biomarker identification is discussed below, organized by cancer type.
Liver
Hepatocarcinoma, or hepatocellular cancer (HCC), has been the focus of metabolic profiling for a number of biomarker discovery and diagnostic models. [48],[49],[50],[51],[52],[53],[54],[55] For example, serum and urine samples from HCC, benign liver tumor, and healthy patients have been analyzed using GC-MS and LC-QTOF-MS to find HCC biomarkers. Around 70 metabolites showed significant differences between cancerous samples and healthy controls. [50] Bile acids, histidine, and inosine levels had large and highly statistically significant changes between normal and HCC samples. Further analysis determined that glycochenodeoxycholic acid, glycocholic acid, taurocholic acid, and chenodeoxycholic acid differed within the HCC samples, showing correlation with liver cirrhosis and hepatitis.
Kidney
Metabolic profiling of urine samples is ideally suited to identify novel markers for early diagnosis of kidney cancer (whose incidence is currently increasing due to lack of an early diagnostic test [56],[57],[58] ) due to the close interaction of urine and the kidneys. [59] Several groups have used metabolomics analysis to distinguish between cancerous and healthy urine samples, finding that acylcarnitines, quinolinate, 4-hydroxybenzoate, and gentisate were differentially accumulated. [58],[60],[61],[62] Serum [38],[63],[64],[65] and tissue [66],[67] samples have also been used to further distinguish between kidney cancer metabolism and normal metabolism.
Breast
Beyond the aforementioned common metabolic alterations in cancer, it has been known for some time that breast cancer cells are also characterized by high levels of total choline containing compounds. [68],[69],[70] This well-known metabolic alteration has served as a gateway for significant metabolomics analysis of breast cancer. [71],[72] Screening for early diagnosis has been shown to be possible using metabolomic analysis of (non-invasive) urine [73] and serum [74],[75],[76] samples, while analysis of breast tissue biopsy samples can be a useful tool as a form of secondary confirmation. For example, HR-MAS-NMR metabolomic analysis of biopsy samples has discriminated between cancerous and normal samples [77] – especially useful as it is non-destructive, allowing other analyses to be performed on the same sample.
Ovarian
A major focus in ovarian cancer metabolomics research has been in early detection, as the 5-year survival rate when caught in early stages is greater than 90%, but when diagnosed in later stages (as it is for most patients) is almost inverted. [57],[78] A number of studies have attempted to use metabolomics analysis of urine or serum as an early diagnosis tool. [39],[79],[80] One particularly promising model used DART-MS to profile the metabolome of 44 ovarian cancer patients and 50 healthy patients through serum samples, obtaining 99% separation accuracy using a customized algorithm. [39]
Colorectal
Diagnostic biomarkers for colorectal cancer (CRC) have also been extensively explored via metabolomics. [81],[82],[83],[84],[85],[86] Metabolic profiling of serum samples from cancer patients and normal controls resulted in the selection of four metabolites (2-hydroxybutyrate, aspartic acid, kynurenine, and cystamine) as the basis of an early diagnostic model. The validated model showed high sensitivity towards detecting early stage CRC. [82] Another study identified approximately 30 marker metabolites with statistically significantly different levels between normal mucosae and CRC tissue samples; most were from pathways expected to be perturbed in cancer, such as glycolysis, lipid metabolism, and nucleotide biosynthesis. [83]
Emerging applications
Metabolomics and metastasis
Metabolomics research has shown promising results for detection of metastasis. Metabolic profiles of serum or urine samples suggest predictive capabilities for diagnosing metastases forming from gastric, [87] CRC, [88] kidney, [64],[67] and breast [74],[75],[89] cancer. Other studies have focused on specific metastatic sites such as leptomeningeal carcinomatosis [90] and bone metastases [91] , the latter of which contain higher levels of cholesterol for prostate cancer metastases when compared to other cancerous bone metastases and normal bone.
Staging of cancer
Beyond detection, metabolomics may also serve a role in distinguishing between different stages of cancer. In one study, GC-MS analysis of serum from pancreatic cancer patients was able to distinguish between Stage III, Stage IVa and Stage IVb groups. [92] Another study used GC-TOF-MS to analyze ovarian cancer samples and showed metabolic distinction of ovarian carcinomas and borderline tumors. [93] In CRC, HR-MAS-NMR profiling not only distinguished between tumor and adjacent mucosa samples, but also between the mucosa samples themselves based on the stage of their adjacent tumor. [94]
Metabolomics and treatment
An emerging field of study for metabolomics is pharmacometabolomics, the use of metabolomics to predict physiological responses for drug efficacy and/or toxicity. There are currently few pharmacometabolomics studies in oncology, but research in the area is expected to grow, [95] particularly since pharmacometabolomics is already achieving widespread attention in other fields [96],[97],[98],[99],[100],[101],[102] . In a pharmacometabolomic study of toxicity effects of capecitabine on CRC patients, lipoprotein-derived lipid levels were discovered to correlate with the intensity of toxicity, yielding predictive capabilities. [103] In another study, metabolic profiling of serum before and during chemotherapy from breast cancer patients with metastasis found that metabolite profiles from human epidermal growth factor receptor 2 (HER2)-positive patients treated with paclitaxel and lapatinib correlated with overall survival and time to progression (though the correlation did not hold across the entire population). [104]
Conclusion and Perspective
Metabolomics holds great promise for advancing the understanding, diagnosis, and treatment of cancer. The approach has already been used to uncover and verify mechanisms of carcinogenesis and proliferation, identify numerous candidate diagnostic biomarkers in biofluid and biopsy samples, and even contribute to the staging of cancers and characterization of treatment efficacy – much of this before metabolomics analysis became more widely accessible to researchers via broader establishment of metabolomics core facilities.
However, some issues must still be better addressed before metabolomics has more broad and direct clinical impact. For example, sample acquisition and preparation must be rigorously standardized in order to produce results that are reproducible enough for clinical applications, since large and fast intracellular changes in metabolite concentrations are possible during biopsies. Moreover, metabolites must be chemically identified, verified, and validated in order to see widespread adoption as clinical diagnostics (even if chemically unidentified metabolites reproducibly distinguish between experimental classes), requiring great strides in development of standard metabolite libraries or de novo chemical identification of unknown metabolites (both active areas of research [105],[106],[107],[108],[109] ). Finally, more widespread clinical trials need to be conducted to provide proper validation for existing (often small sample size) biomarker research.
Future research frontiers in cancer metabolomics offer great promise, though with significant challenges. An obvious goal is to translate metabolomics measurements into deeper biological understanding of the condition, ultimately enabling better drug design and development. An increasingly popular approach to this is through the integration of multiple “omics” fields. [110],[111] Integration of, for example, transcriptomic and metabolomic data has enabled deeper analysis of chemosensitive pathways [112] and breast cancer [113] , and may provide further validation and understanding (and thus potential clinical applications) of discoveries.
Another prominent goal is identification of biomarkers specifically targeted toward early diagnosis. Detecting early-stage cancer, where survival rates and treatment efficacy are vastly improved, would have a transformative impact on cancer diagnosis and treatment. It remains to be seen, though, whether metabolic changes will be strong enough early indicators to be detected through non-invasive biofluid samples, or even through targeted biopsies.
Finally, the interpretation of biofluid diagnostics may prove particularly difficult. While existing analysis of blood or urine samples may find analytes that distinguish cancerous from non-cancerous samples, the metabolites detected are often “generic” cancer-associated metabolites and may not distinguish, for example, kidney cancer from liver cancer. The commonality of dysfunctional metabolism across all types of cancer may in this case turn out to be a hindrance to interpretability and direct diagnosis; more work is needed to assess how useful such non-invasive tests could be.
Challenges notwithstanding, metabolomics is a field rife with promise to help decrease the burden and impact of all types of cancer on society.
Acknowledgments
The authors acknowledge support from DARPA grant YFA N66001-11-1-4128, NIH grant R21 CA167500 and the Integrative Bio Systems Institute of Georgia Tech.
References
| 1. | Assfalg M, Bertini I, Colangiuli D, Luchinat C, Schäfer H, Schütz B, et al. Evidence of different metabolic phenotypes in humans. Proc Natl Acad Sci U S A 2008;105:1420-4.  |
| 2. | Bernini P, Bertini I, Luchinat C, Nepi S, Saccenti E, Schäfer H, et al. Individual human phenotypes in metabolic space and time. J Proteome Res 2009;8:4264-71.  |
| 3. | Fiehn O. Metabolomics: The link between genotypes and phenotypes. Plant Mol Biol 2002;48:155-71.  |
| 4. | O’Connell TM. Recent advances in metabolomics in oncology. Bioanalysis 2012;4:431-51.  |
| 5. | Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011;144:646-74.  |
| 6. | Warburg O, Posener K, Negelein E. On the metabolism of carcinoma cells. Biochem Z 1924;152:309-44.  |
| 7. | Warburg O. On the origin of cancer cells. Science 1956;123:309-14.  |
| 8. | Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011;11:85-9.  |
| 9. | Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 2004;64:3892-9.  |
| 10. | Semenza GL. HIF-1: Upstream and downstream of cancer metabolism. Curr Opin Genet Dev 2010;20:51-6.  |
| 11. | Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 2006;3:187-97.  |
| 12. | Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab 2006;3:177-85.  |
| 13. | Lu CW, Lin SC, Chen KF, Lai YY, Tsai SJ. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J Biol Chem 2008;283:28106-14.  |
| 14. | Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem 1997;272:22776-80.  |
| 15. | Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science 2006;312:1650-3.  |
| 16. | Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006;126:107-20.  |
| 17. | Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res 2005;65:177-85.  |
| 18. | Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005;7:77-85.  |
| 19. | Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell 2005;8:143-53.  |
| 20. | King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 2006;25:4675-82.  |
| 21. | Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet 2005;14:2231-9.  |
| 22. | MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol 2007;27:3282-9.  |
| 23. | DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A 2007;104:19345-50.  |
| 24. | Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer 2008;8:51-6.  |
| 25. | Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res 2010;70:859-62.  |
| 26. | Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009;458:762-5.  |
| 27. | Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008;452:230-3.  |
| 28. | Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009;324:1029-33.  |
| 29. | Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol 2011;13:310-6.  |
| 30. | Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 2010;329:1492-9.  |
| 31. | Dunn WB, Bailey NJ, Johnson HE. Measuring the metabolome: Current analytical technologies. Analyst 2005;130:606-25.  |
| 32. | Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst 2012;137:293-300.  |
| 33. | Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 2011;40:387-426.  |
| 34. | Duarte IF, Gil AM. Metabolic signatures of cancer unveiled by NMR spectroscopy of human biofluids. Prog Nucl Magn Reson Spectrosc 2012;62:51-74.  |
| 35. | Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc 2010;5:1019-32.  |
| 36. | Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab 2012;15:827-37.  |
| 37. | Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia 2011;13:81-97.  |
| 38. | Lin L, Yu Q, Yan X, Hang W, Zheng J, Xing J, et al. Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst 2010;135:2970-8.  |
| 39. | Zhou M, Guan W, Walker LD, Mezencev R, Benigno BB, Gray A, et al. Rapid mass spectrometric metabolic profiling of blood sera detects ovarian cancer with high accuracy. Cancer Epidemiol Biomarkers Prev 2010;19:2262-71.  |
| 40. | Blekherman G, Laubenbacher R, Cortes DF, Mendes P, Torti FM, Akman S, et al. Bioinformatics tools for cancer metabolomics. Metabolomics 2011;7:329-43.  |
| 41. | Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73.  |
| 42. | Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739-44.  |
| 43. | Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21:297-308.  |
| 44. | Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A 2011;108:3270-5.  |
| 45. | Jentzmik F, Stephan C, Miller K, Schrader M, Erbersdobler A, Kristiansen G, et al. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. Eur Urol 2010;58:12-8.  |
| 46. | Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009;457:910-4.  |
| 47. | Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012;336:1040-4.  |
| 48. | Yang J, Xu G, Zheng Y, Kong H, Pang T, Lv S, et al. Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitis and hepatocirrhosis diseases. J Chromatogr B Analyt Technol Biomed Life Sci 2004;813:59-65.  |
| 49. | Wang X, Zhang A, Sun H, Yan G, Han Y, Ye Y. Urinary metabolic profiling identifies a key role for glycocholic acid in human liver cancer by ultra-performance liquid-chromatography coupled with high-definition mass spectrometry. Clin Chim Acta 2013;418:86-90.  |
| 50. | Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics 2011;10:M110.004945.  |
| 51. | Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, et al. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci 2009;100:782-5.  |
| 52. | Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res 2011;71:6590-600.  |
| 53. | Shariff MI, Gomaa AI, Cox IJ, Patel M, Williams HR, Crossey MM, et al. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: A validation study. J Proteome Res 2011;10:1828-36.  |
| 54. | Xue R, Lin Z, Deng C, Dong L, Liu T, Wang J, et al. A serum metabolomic investigation on hepatocellular carcinoma patients by chemical derivatization followed by gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom 2008;22:3061-8.  |
| 55. | Yang Y, Li C, Nie X, Feng X, Chen W, Yue Y, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res 2007;6:2605-14.  |
| 56. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300.  |
| 57. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10-29.  |
| 58. | Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol Oncol 2011;29:551-7.  |
| 59. | Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, et al. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics 2009;8:558-70.  |
| 60. | Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal Biochem 2007;363:185-95.  |
| 61. | Kim K, Taylor SL, Ganti S, Guo L, Osier MV, Weiss RH. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS 2011;15:293-30.  |
| 62. | Ganti S, Taylor SL, Kim K, Hoppel CL, Guo L, Yang J, et al. Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer 2012;130:2791-800.  |
| 63. | Lin L, Huang Z, Gao Y, Yan X, Xing J, Hang W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J Proteome Res 2011;10:1396-405.  |
| 64. | Gao H, Dong B, Liu X, Xuan H, Huang Y, Lin D. Metabonomic profiling of renal cell carcinoma: High-resolution proton nuclear magnetic resonance spectroscopy of human serum with multivariate data analysis. Anal Chim Acta 2008;624:269-77.  |
| 65. | Zira AN, Theocharis SE, Mitropoulos D, Migdalis V, Mikros E. (1) H NMR metabonomic analysis in renal cell carcinoma: A possible diagnostic tool. J Proteome Res 2010;9:4038-44.  |
| 66. | Catchpole G, Platzer A, Weikert C, Kempkensteffen C, Johannsen M, Krause H, et al. Metabolic profiling reveals key metabolic features of renal cell carcinoma. J Cell Mol Med 2011;15:109-18.  |
| 67. | Gao H, Dong B, Jia J, Zhu H, Diao C, Yan Z, et al. Application of ex vivo (1) H NMR metabonomics to the characterization and possible detection of renal cell carcinoma metastases. J Cancer Res Clin Oncol 2012;138:753-61.  |
| 68. | Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res 1999;59:80-4.  |
| 69. | Katz-Brull R, Seger D, Rivenson-Segal D, Rushkin E, Degani H. Metabolic markers of breast cancer: Enhanced choline metabolism and reduced choline-ether-phospholipid synthesis. Cancer Res 2002;62:1966-70.  |
| 70. | Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res 2004;64:4270-6.  |
| 71. | Oakman C, Tenori L, Biganzoli L, Santarpia L, Cappadona S, Luchinat C, et al. Uncovering the metabolomic fingerprint of breast cancer. Int J Biochem Cell Biol 2011;43:1010-20.  |
| 72. | Oakman C, Tenori L, Cappadona S S, Luchinat C, Bertini I, Leo A. Targeting Metabolomics in Breast Cancer. Curr Breast Cancer Rep 2012;4:249-56.  |
| 73. | Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res 2010;16:5835-41.  |
| 74. | Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, et al. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res 2010;70:8309-18.  |
| 75. | Oakman C, Tenori L, Claudino WM, Cappadona S, Nepi S, Battaglia A, et al. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol 2011;22:1295-301.  |
| 76. | Gu H, Pan Z, Xi B, Asiago V, Musselman B, Raftery D. Principal component directed partial least squares analysis for combining nuclear magnetic resonance and mass spectrometry data in metabolomics: Application to the detection of breast cancer. Anal Chim Acta 2011;686:57-63.  |
| 77. | Li M, Song Y, Cho N, Chang JM, Koo HR, Yi A, et al. An HR-MAS MR metabolomics study on breast tissues obtained with core needle biopsy. PLoS One 2011;6:e25563.  |
| 78. | Cho KR, Shih IeM. Ovarian cancer. Annu Rev Pathol 2009;4:287-313.  |
| 79. | Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, et al. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer 2005;113:782-8.  |
| 80. | Zhang T, Wu X, Ke C, Yin M, Li Z, Fan L, et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J Proteome Res 2013;12:505-12.  |
| 81. | Yue H, Wang Y, Zhang Y, Ren H, Wu J, Ma L, et al. A Metabonomics Study of Colorectal Cancer by RRLC-QTOF/MS. J Liq Chromatogr Relat Technol 2013;36:428-38.  |
| 82. | Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One 2012;7:e40459.  |
| 83. | Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J Proteome Res 2009;8:352-61.  |
| 84. | Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res 2009;8:4844-50.  |
| 85. | Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, et al. Urinary metabonomic study on colorectal cancer. J Proteome Res 2010;9:1627-34.  |
| 86. | Li S, Guo B, Song J, Deng X, Cong Y, Li P, et al. Plasma choline-containing phospholipids: potential biomarkers for colorectal cancer progression. Metabolomics 2013;9:202-12.  |
| 87. | Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, et al. Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol 2011;17:727-34.  |
| 88. | Bertini I, Cacciatore S, Jensen BV, Schou JV, Johansen JS, Kruhøffer M, et al. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res 2012;72:356-64.  |
| 89. | Lu X, Bennet B, Mu E, Rabinowitz J, Kang Y. Metabolomic changes accompanying transformation and acquisition of metastatic potential in a syngeneic mouse mammary tumor model. J Biol Chem 2010;285:9317-21.  |
| 90. | Cho HR, Wen H, Ryu YJ, An YJ, Kim HC, Moon WK, et al. An NMR metabolomics approach for the diagnosis of leptomeningeal carcinomatosis. Cancer Res 2012;72:5179-87.  |
| 91. | Thysell E, Surowiec I, Hörnberg E, Crnalic S, Widmark A, Johansson AI, et al. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. PLoS One 2010;5:e14175.  |
| 92. | Nishiumi S, Shinohara M, Ikeda A, Yoshie T, Hatano N, Kakuyama S, et al. Serum metabolomics as a novel diagnostic approach for pancreatic cancer. Metabolomics 2010;6:518-28.  |
| 93. | Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res 2006;66:10795-804.  |
| 94. | Jimenez B, Mirnezami R, Kinross JM, Cloarec O, Keun HC, Holmes E, et al.(1) H HR-MAS NMR Spectroscopy of Tumor Induced Local Metabolic “Field-Effects” enables Colorectal Cancer Staging and Prognostication. J Proteome Res 2012;12:959-68.  |
| 95. | Corona G, Rizzolio F, Giordano A, Toffoli G. Pharmaco-metabolomics: An emerging “omics” tool for the personalization of anticancer treatments and identification of new valuable therapeutic targets. J Cell Physiol 2012;227:2827-31.  |
| 96. | Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006;440:1073-7.  |
| 97. | Li H, Ni Y, Su M, Qiu Y, Zhou M, Qiu M, et al. Pharmacometabonomic phenotyping reveals different responses to xenobiotic intervention in rats. J Proteome Res 2007;6:1364-70.  |
| 98. | Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 2009;106:14728-33.  |
| 99. | Patterson AD, Slanar O, Krausz KW, Li F, Höfer CC, Perlík F, et al. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J Proteome Res 2009;8:4293-300.  |
| 100. | West PR, Weir AM, Smith AM, Donley EL, Cezar GG. Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol Appl Pharmacol 2010;247:18-27.  |
| 101. | Nicholson JK, Wilson ID, Lindon JC. Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics 2011;12:103-11.  |
| 102. | Abo R, Hebbring S, Ji Y, Zhu H, Zeng ZB, Batzler A, et al. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes’ single-nucleotide polymorphism imputation: Selective serotonin reuptake inhibitor response pharmacogenomics. Pharmacogenet Genomics 2012;22:247-53.  |
| 103. | Backshall A, Sharma R, Clarke SJ, Keun HC. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin Cancer Res 2011;17:3019-28.  |
| 104. | Tenori L, Oakman C, Claudino WM, Bernini P, Cappadona S, Nepi S, et al. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: A pilot study. Mol Oncol 2012;6:437-44.  |
| 105. | Hummel J, Selbig J, Walther D, Kopka J. The Golm Metabolome Database: a database for GC-MS based metabolite profiling. In: Nielsen J, Jewett M, editors. Metabolomics. 1 ed. Leipzig, Germany: Springer Berlin Heidelberg; 2007. p. 75-95.  |
| 106. | Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0: The Human Metabolome Database in 2013. Nucleic Acids Res 2013;41:D801-7.  |
| 107. | Ludwig C, Easton JM, Lodi A, Tiziani S, Manzoor SE, Southam AD, et al. Birmingham Metabolite Library: a publicly accessible database of 1-D H-1 and 2-D H-1 J-resolved NMR spectra of authentic metabolite standards (BML-NMR). Metabolomics 2012;8:8-18.  |
| 108. | Kind T, Wohlgemuth G, Lee do Y, Lu Y, Palazoglu M, Shahbaz S, et al. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 2009;81:10038-48.  |
| 109. | Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics 2007;8:105.  |
| 110. | Benjamin DI, Cravatt BF, Nomura DK. Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab 2012;16:565-77.  |
| 111. | Casado-Vela J, Cebrián A, Gómez del Pulgar MT, Lacal JC. Approaches for the study of cancer: Towards the integration of genomics, proteomics and metabolomics. Clin Transl Oncol 2011;13:617-28.  |
| 112. | Cavill R, Kamburov A, Ellis JK, Athersuch TJ, Blagrove MS, Herwig R, et al. Consensus-phenotype integration of transcriptomic and metabolomic data implies a role for metabolism in the chemosensitivity of tumour cells. PLoS Comput Biol 2011;7:e1001113.  |
| 113. | Borgan E, Sitter B, Lingjærde OC, Johnsen H, Lundgren S, Bathen TF, et al. Merging transcriptomics and metabolomics: Advances in breast cancer profiling. BMC Cancer 2010;10:628.  |
Authors
Kathleen A. Vermeersch: School of Chemical & Biomolecular Engineering and Institute for Bioengineering & Bioscience, Georgia Institute of Technology, 311 Ferst Dr., NW Atlanta, GA 30332-0100, USA
Dr. Mark P. Styczynski: School of Chemical & Biomolecular Engineering and Institute for Bioengineering & Bioscience, Georgia Institute of Technology, 311 Ferst Dr., NW Atlanta, GA 30332-0100, USA
